Using IGV via OnDemand¶
The Integrative Genomics Viewer (IGV) is a high-performance, easy-to-use, interactive tool for the visual exploration of genomic data. It supports flexible integration of all the common types of genomic data and metadata, investigator-generated or publicly available, loaded from local or cloud sources. Using IGV on Apocrita via OnDemand allows you to run a GUI interface. Please refer to the overview section for instructions on how to login to OnDemand.
Starting an IGV session¶
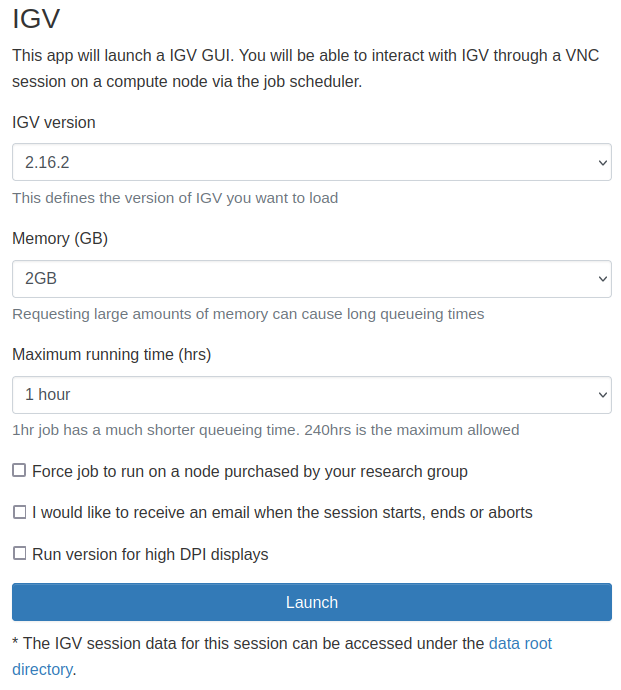
Select IGV from the GUIs list of the Interactive Apps drop-down menu, or from the My Interactive Sessions page.
Choose the resources your job will need. Choosing a 1 hour maximum running time is the best option for getting a session quickly, unless you have access to owned nodes which may offer sessions immediately for up to 24hrs if resources are available.
High DPI displays
If your application interface is very small (you have high DPI display), you
may tick the Run version for high DPI displays option to run the high DPI
version, adapted for such displays.
Once clicking Launch, the request will be queued, and when resources have been allocated, you will be presented with the option to connect to the session by clicking on the blue Launch IGV button.
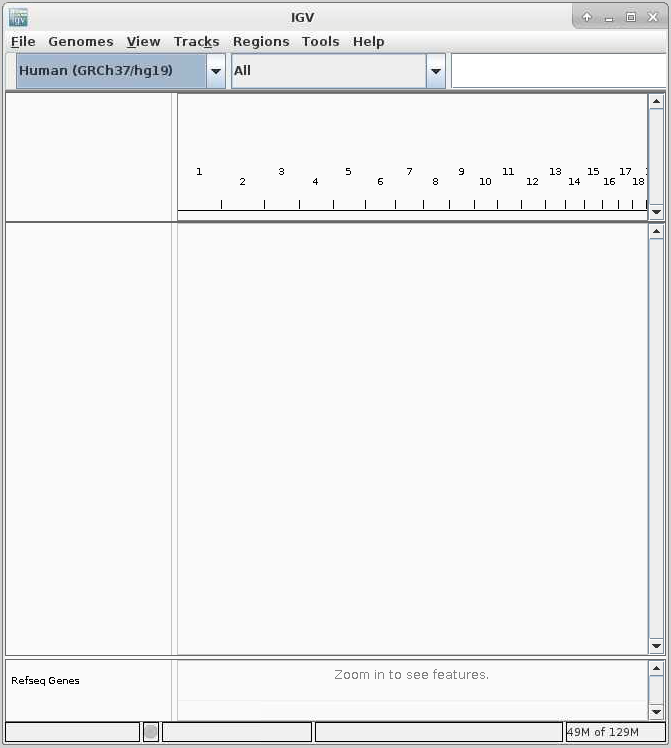
Once connected, the familiar IGV interface is presented, and you will be able to use the allocated resources, and access your research data located on Apocrita.
Exiting the session¶
If a session exceeds the requested running time, it will be killed. You can finish your session using one of the following methods:
- clicking [x] in the upper right corner of the IGV application
- selecting File -> Exit from the application menus
- clicking the red Delete button for the relevant session on the My Interactive Sessions page.
After a session ends, the resources will be returned to the cluster queues.